A graph-based approach to spike protein S of SARS-CoV-2
A research team of PhD and graduate students from Bondar's research group designed, implemented and applied a graph-based approach to investigate spike protein S of SARS-CoV-2. They identified dynamic hydrogen-bond communication networks in spike protein.
News from Dec 10, 2020
The surface of the SARS-CoV-2 virion particle is decorated with proteins denoted as spike proteins S, which bind to ACE2 receptors of the host cell. Description of the mechanism by which particular conformations of protein S are selected for binding to the receptor could guide the development of peptides to hinder binding.
Computational approaches have the potential to provide a detailed molecular picture of interactions that govern the binding of protein S to the host receptor, however the large size of protein S makes it difficult to evaluate the relative importance of specific regions or groups of the protein. To tackle this challenge, the paper by Karathanou and colleagues reports a graph-based approach that enables efficient analyses of the structural dynamics of protein S in terms of hydrogen bonding.
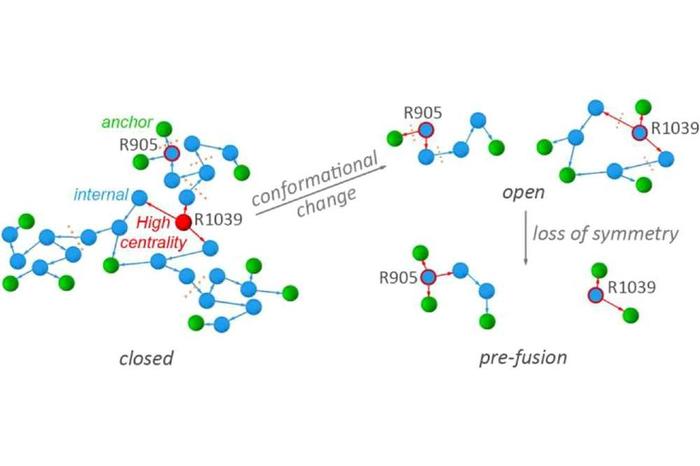
With this approach, Karathanou and colleagues found that the conformational transition of protein S from a closed conformation to a pre-fusion conformation that can bind to protein S associates with loss of three-fold compositional symmetry of hydrogen-bond clusters located at key sites of protein S. A complex, extended network of hydrogen bonds was identified at the binding interface between protein S and ACE2. In this network, amino acid residue N501 of protein S has a central role. Intriguingly, N501 is among the three groups mutated in the new 501Y.V2 SARS-CoV-2 lineage (Tegally et al, medRxiv, https://doi.org.10.1101/2020).
The research reported by Karathanou and colleagues involved the joined effort of Freie Universität doctoral students Konstantina Karathanou, Michalis Lazaratos, and Krzysztof Buzar, Master student Eva Bertalan, student assistant Malte Siemers. The study has been published in the Journal of Structural Biology.
Keywords
- ACE2
- basic research
- Bimolecular structure
- biophysics
- Conformational plasticity
- Dynamic hydrogen-bond clusters
- fundamental research
- Hydrogen bonding
- SARS-CoV-2 protein S