Research
Last update: 2020
Our research aims at understanding chemical and physical processes in biological macromolecules such as proteins and enzymes by means of computer simulations. We are targeting the rational development of biomimetic substitutes for enzymes (catalysts), the design of specific inhibitors and rational protein engineering, using molecular dynamics simulations and combined QM/MM approaches. Current applications are protein-substrate interaction and substrate recognition, exploring enzymatic reaction pathways and enzymatic catalysis as well as photoinduced transformations from conformational transitions to reactions.
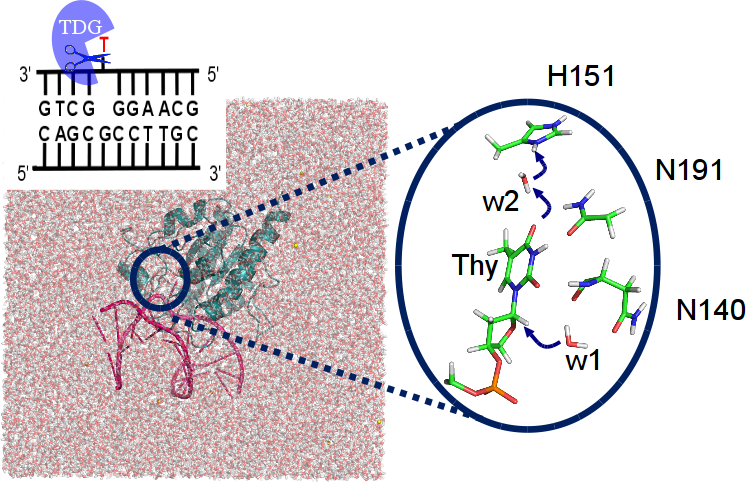 |
Our DNA is constantly threatened by damages due to e.g. UV radiation or chemical agents. One such damage, methyl-cytosine deamination results in a thymine base, forming a mismatch with the guanine base on the complementary strand that could lead to mutations and genetic deseases if translated. Thymine DNA Glycosylase (TDG) is an enzyme of the base excision repair system, a machinery to correct such and other DNA damges. It likely recognises the target thymine by the different conformational dynamics of the mismatched DNA (paper) . For another form of methyl-cytosine damage (oxidations), this mode of recognition could, however, not be confirmed (paper). Our computations of the chemical step of base excision by TDG, i.e. removal of the mismatched thymine, elucidate the atomistic details of the reaction mechanism (paper). |
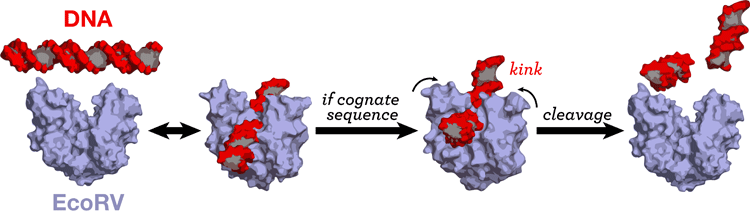 |
Restriction enzymes cleave DNA, highly specifically at a particular sequence. EcoRV induces a strong helix kink to only its cognate sequence. We are exploring the origins of the sequence specificity in EcoRV by all-atom Molecular Dynamics simulations of protein-DNA complexes with different sequences, the cognate and non-cognate sequences. (paper) MD simulations have also enabled us to elucidate the role of the metal ions in DNA recognition (paper). |
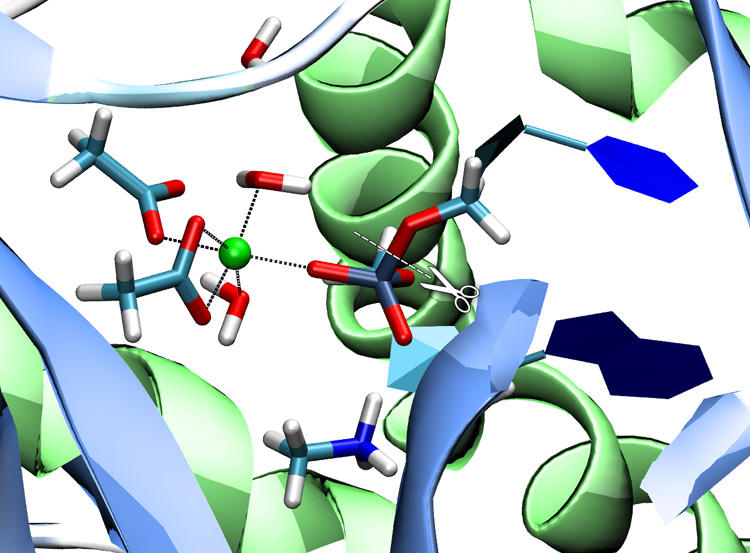 |
Phosphate transfer reactions are essential to many biochemical processes including nucleotide synthesis, signal transduction, and cell growth regulation. Despite their stability against hydrolysis, phosphate esters can be processed by phosphate transferring enzymes under mild (physiological) conditions in minutes or even seconds. We are investigating the reaction mechanisms of a variety of phosphate transferring enzymes (nucleases, kinases, phosphatases) applying QM/MM methods. (paper) |
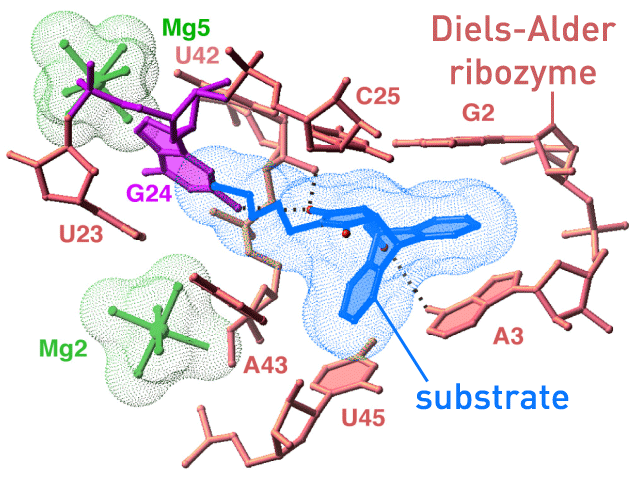 |
Biochemical reactions are not exclusively accelerated by protein enzymes, Ribonucleic Acid, RNA, can also act as catalyst. Andres Jäschke and coworkers have synthesized an artificial Diels-Alderase. We are working together on elucidating the enzymatic reaction mechanism and the reasos for the high speciticity of the ribozyme. (paper1/paper2) |
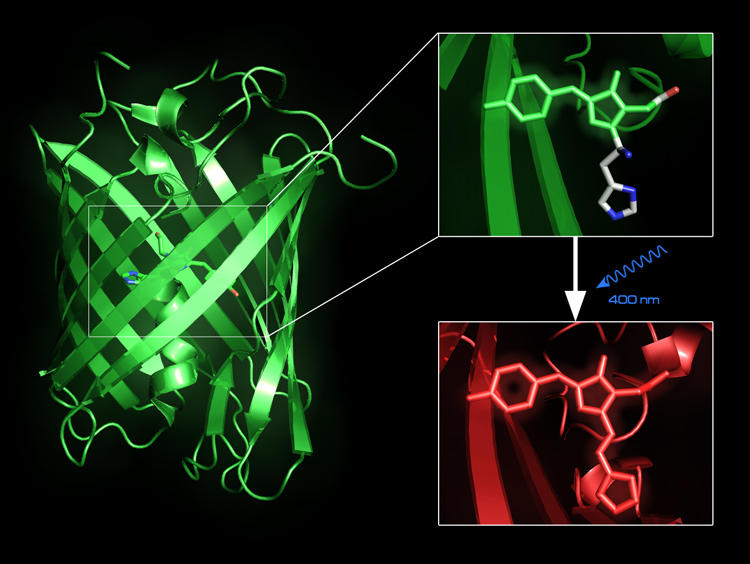 |
Fluorescent proteins, such as the Green Fluorescent Protein, can be used to trace intracellular processes in vivo. Recently, a new fluorescent protein, EosFP, has been designed, which upon irradiation with blue light changes its emission from green to red light whose wavelength does not overlap with the green autofluorescence of living cells. We are conducting simulation studies in order to understand the relationship between structure and fluorescent properties.(paper) |
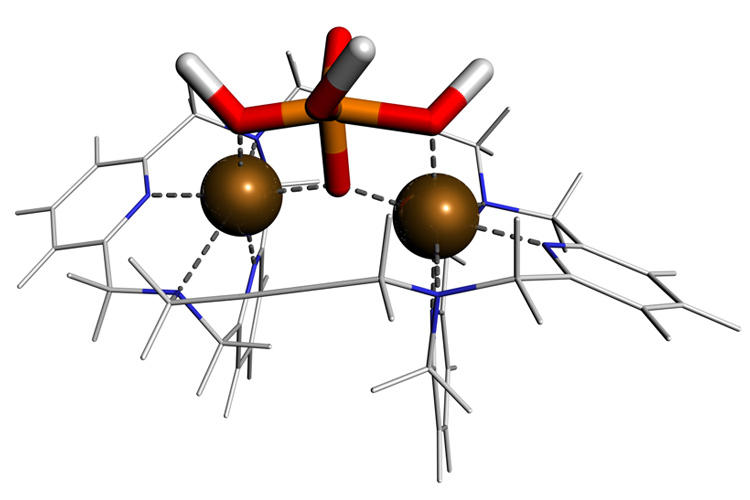 |
Based on the knowledge from studying enzymatic pathways we are working on the rational design of bio-mimetic catalysts for phosphoester hydrolysis reactions. In collaboration with Roland Krämer we have characterised a synthetic catalyst for phosphate transfer by computer simulation, revealing a reaction mechanism similar to that of the exonuclease enzyme. (paper) |